How Ikigai Labs Serves Interactive AI Workflows at Scale using Ray Serve
LinkWhat We Do at Ikigai Labs
In short, Ikigai Labs provides AI-charged spreadsheets: an AI augmented data processing and analytics collaborative, cloud platform that can be used with an ease of spreadsheet. Within the platform, powerful AI enabled end-to-end automated workflows can be built while staying in the comfort of a spreadsheet. The Ikigai Labs platform offers a collaborative space that brings data integration, processing, visualization, and interactive dashboards to users at unprecedented ease and at scale. Users can utilize the platform to automate, maintain, and enhance day-to-day mission critical operations.
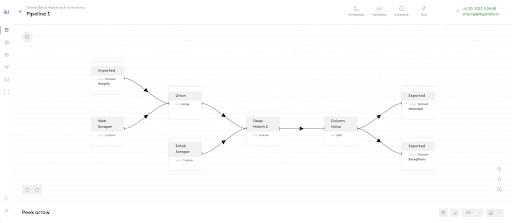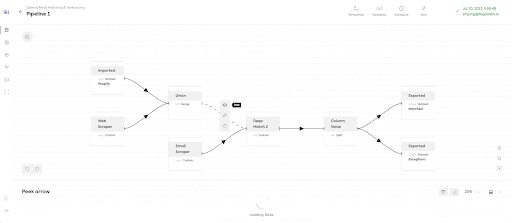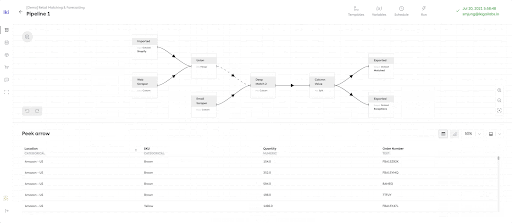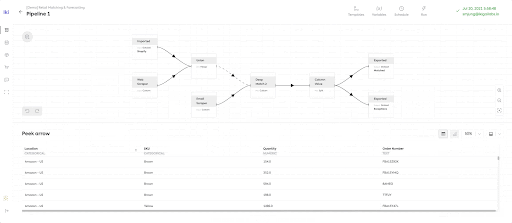
While the platform supports various features, they all revolve around the data processing pipeline. The Ikigai data pipeline encapsulates many components such as data wrangling steps, data connectors, predictive functionality, what-if scenario analysis and dashboards. Making the data pipeline scalable and highly interactive by enabling users to inject custom Python code into a traditional data pipelining platform involved solving many technical challenges. This post dives into these challenges and how Ray and Ray Serve provided excellent flexibility in resolving them.
LinkIkigai Data Pipeline on Ray
The Ikigai data pipeline aims to achieve the following challenging goals simultaneously:
Mission-Critical data pipelines to address any real-world use case of data-centric tasks. By allowing end users to inject custom Python code on top of traditional data wrangling and preparation features, the data pipeline can adapt to any task requirements.
Highly Interactive data pipelines to bring transparency in every step of the data pipeline. End users can interact with their datasets at any point with a simple spreadsheet or Jupyter notebook view.
Instantly Browsable data pipelines to enable sub-second data browsing of potentially extremely large datasets. The Ikigai data platform offers a unique way of viewing the intermediate state of datasets in the middle of the pipeline, called ‘peeking’.
Most existing data platforms provide either interactive experience but for small datasets (e.g. traditional spreadsheets) or ability to deal with large datasets but in an “offline” or “batch processing” environment (e.g. Apache Spark). At Ikigai, we need the data pipeline to achieve data interactivity while maintaining massive scalability to accomplish the above three missions. This is what makes engineering at Ikigai Labs extremely challenging. At Ikigai Labs, we overcame this challenge by developing a novel computational and data processing architecture with the help of AI. This architecture is made operational using a distributed execution engine: Ray
LinkScalability with Ray Core
Ray is an open source project which provides a simple and universal API for building distributed applications with Python as a first-class citizen. It made it easier for small teams of data engineers and data scientists like ourselves to scale Python-based applications into distributed applications with minimal code changes.
1import pandas as pd
2
3# Business Logics
4def deduplicate(df):
5 df.drop_duplicates(inplace=True)
6 return df
7
8def sort(df, target_columns, descending):
9 df.sort_values(
10 target_columns,
11 ascending=not descending,
12 inplace=True
13 )
14 return data
15
16df = pd.DataFrame([[1, 2, 3], [1, 2, 3], [2, 3, 4]])
17df = deduplicate(df=df)to
1import pandas as pd
2import ray
3
4ray.init()
5
6# Business Logics stay unchanged
7def deduplicate(df):
8 df.drop_duplicates(inplace=True)
9 return df
10
11def sort(df, target_columns, descending):
12 df.sort_values(
13 target_columns,
14 ascending=not descending,
15 inplace=True
16 )
17 return df
18
19df = pd.DataFrame([[1, 2, 3], [1, 2, 3], [2, 3, 4]])
20obj_ref = ray.remote(deduplicate).remote(df=df)
21result_df = ray.get(df_obj_ref)This shows that we were able to adopt Ray Core into our users’ custom Python codebase and scale it without any modification to their code. With Ray Core, unlike frameworks like Spark, users do not need to formulate their codebase into a specific format and understand the complex dependencies to massively scale their data pipeline. While Ray Core can transform any arbitrary Python code from a user into a parallelized distributed application, we focused on a library in the Ray ecosystem to achieve data interactivity as well: Ray Serve.
LinkInteractivity with Ray Serve
Ray Serve is an easy-to-use and scalable model serving library built on Ray. Being framework-agnostic, Ray Serve can serve not only the various deep learning models, but also arbitrary Python code in a distributed manner. Since one of the biggest missions in the Ikigai data pipeline is to run user’s arbitrary Python code at scale with interactivity, Ray Serve provided answers to many challenges we faced as it enabled us to serve users’ code with real-time interaction.
LinkIkigai + Ray Serve: Resolving the Challenges We Faced
As the Ikigai data pipeline aims to run arbitrary Python code at scale with data interactivity, we faced a few challenges coming from the nature of arbitrary code.
LinkConflicting Python Library Dependencies
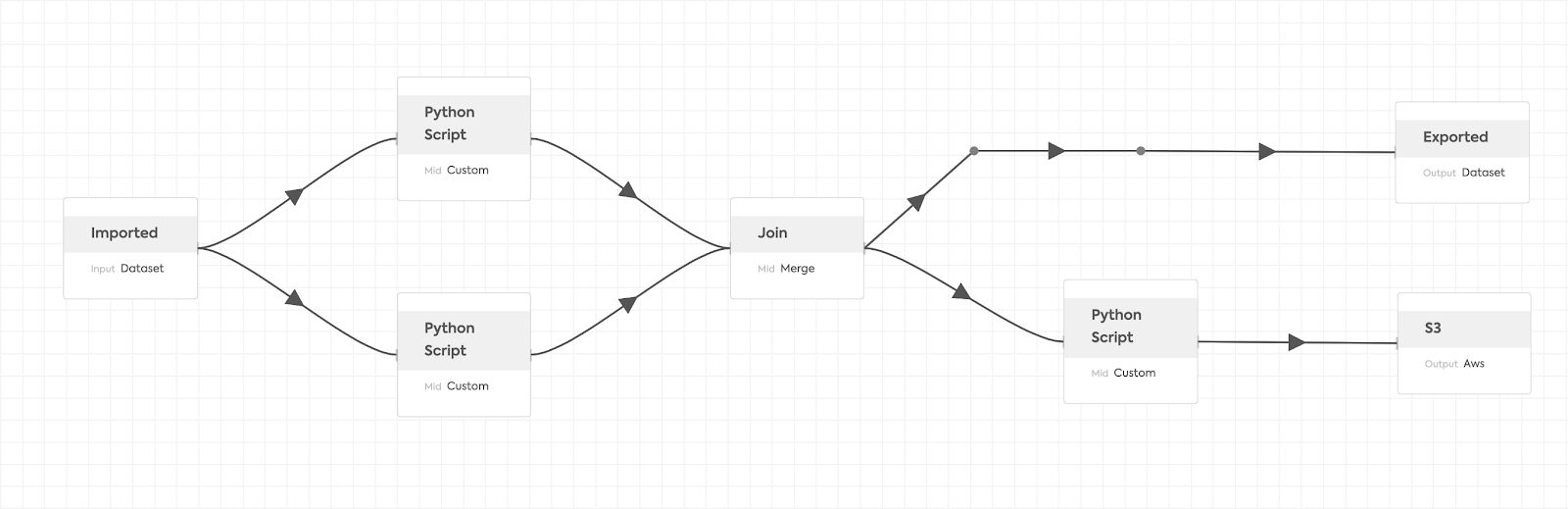
In the Ikigai data pipeline, it is common to have multiple custom Python scripts injected into a single pipeline, where each script requires different Python libraries to be installed. This becomes an issue when the requirements of different scripts start conflicting with each other. Such issues become even more complicated when the data pipeline operates on a distributed cluster as it can be a nightmare to manage different Python environments even within a single machine setup.
As an answer to the challenge, we decided to serve custom Python scripts with Ray Serve. When defining a Serve deployment, Ray Serve lets you define a customizable Conda environment manifest, which will be automatically installed and managed in the Ray cluster. Custom Python scripts are assigned to Serve deployments based on their dependencies, so the data pipeline can handle as many custom Python scripts as needed without worrying about conflicts.
1import ray
2from ray import serve
3
4ray.init(address="auto")
5
6conda_env = {
7 "channels": ["conda-forge", "defaults"],
8 "dependencies": [
9 "python=3.7",
10 {
11 "pip": [
12 "numpy==1.20.2",
13 "pandas==1.2.4",
14 "scipy==1.6.3",
15 "boto3==1.7.0"
16 ]
17 }
18 ]
19}
20
21@serve.deployment(
22 name="custom_service",
23 ray_actor_options={"runtime_env": {"conda": conda_env}}
24)
25class CustomService:
26
27 def deduplicate(self, data):
28 data.drop_duplicates(inplace=True)
29 return data
30
31 def sort(self, data, target_columns, descending):
32 data.sort_values(
33 target_columns,
34 ascending=not descending,
35 inplace=True
36 )
37 return data
38
39CustomService.deploy()The definition of the Python environment manifest will be fed into the serve.deployment decorator, making Conda environment management very lightweight.
LinkTask Overhead for ‘Peek’
To achieve the mission of ‘instantly browsable’ data pipeline, the Ikigai data pipeline enabled the sub-second peeking of intermediate datasets.
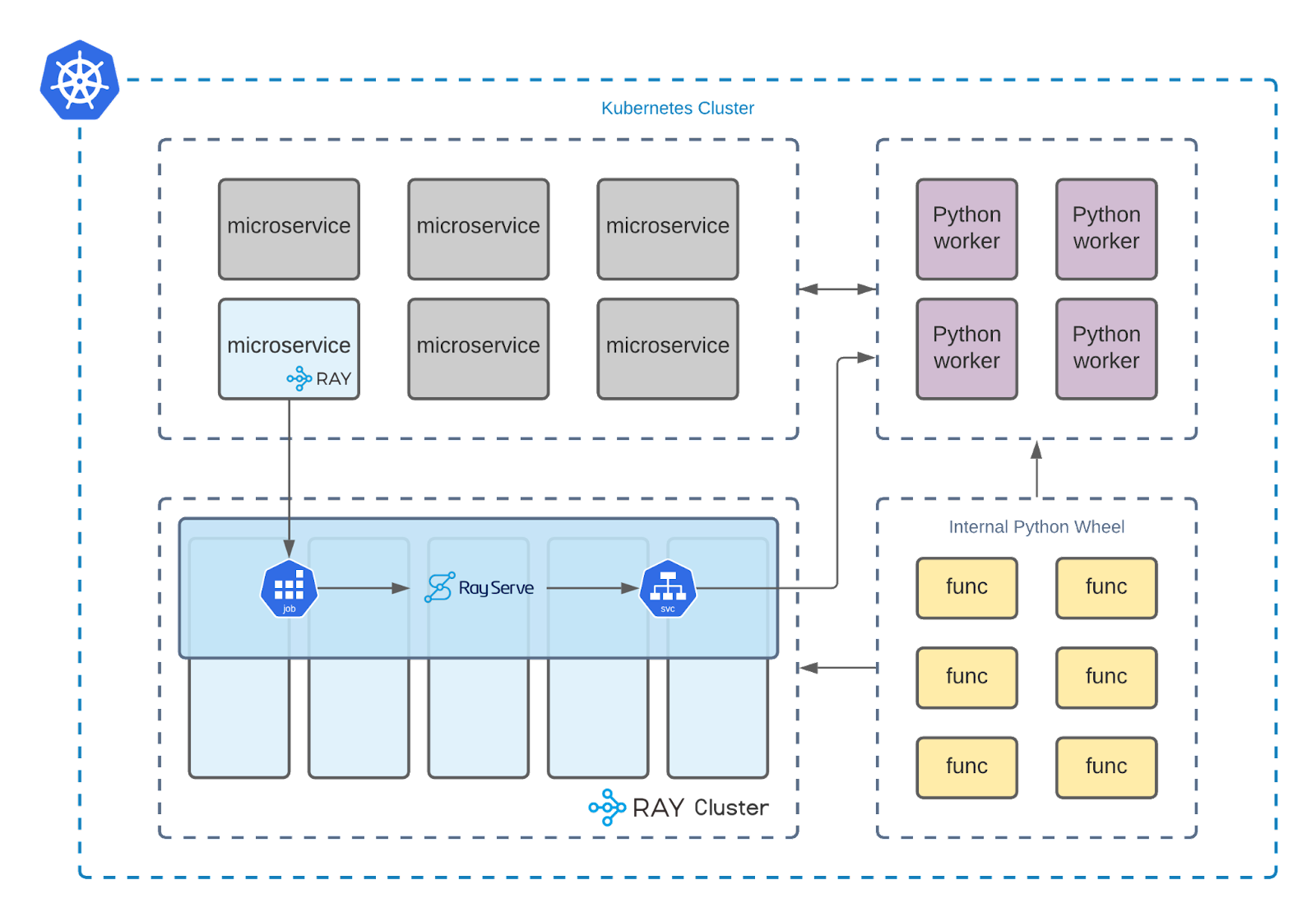
In the data pipeline cluster, there exist internal Python libraries which hold all the traditional data preparation functions. Ray cluster and simple Python runner containers utilize the same functionalities in different locations, depending on the type of pipeline task: scalable run or peek run. In the scenario of scalable run, tasks are run on Ray cluster, utilizing Ray’s distributed computing engine.
On the other hand, the data pipeline will simply utilize local Python runner containers for peek runs. This design decision was made to eliminate the ‘task submission overhead’ (about ~10 seconds) which is generated when submitting a Python task definition to a Ray Cluster. With such a design, peek runs on local Python runner containers that process subsets of the whole dataset in under a second as they don’t submit any tasks to the Ray cluster.
Here, a specific challenge emerges because we serve users’ custom Python scripts with Ray Serve. Ray Serve’s dependency management solution is essential in this case but now peek runs will suffer from the task submission overhead time problem. Thus, in order to leverage Ray Serve without incurring the task submission overhead we looked into additional features. Thankfully, Ray Serve has adopted FastAPI to expose HTTP endpoints for Serve instances which allows us to peek with Ray Serve without incurring additional overhead.
1@serve.deployment(name="custom_service")
2class CustomService:
3
4 def deduplicate(self, data):
5 data.drop_duplicates(inplace=True)
6 return datato
1app = FastAPI()
2
3@serve.deployment(name="custom_service")
4@serve.ingress(app)
5class CustomService:
6 @app.post("/deduplicate")
7 def deduplicate(self, request: typing.Dict):
8 data = request["data"]
9 columns = request["columns"]
10
11 import pandas as pd
12 data = pd.DataFrame(data, columns=columns)
13
14 data.drop_duplicates(inplace=True)
15 return data.values.tolist()Having this easy transformation of a Serve instance into a HTTP compatible version, simple Python runner containers are able to run custom Python scripts without suffering from the task submission overhead.
1import requests
2
3df = requests.post(
4 "http://ray-serve-service.ray:8000/custom_service/deduplicate",
5 json={"data": [[1, 2, 3], [1, 2, 3], [2, 3, 4]]}
6).json()LinkHandling Concurrent Deployments
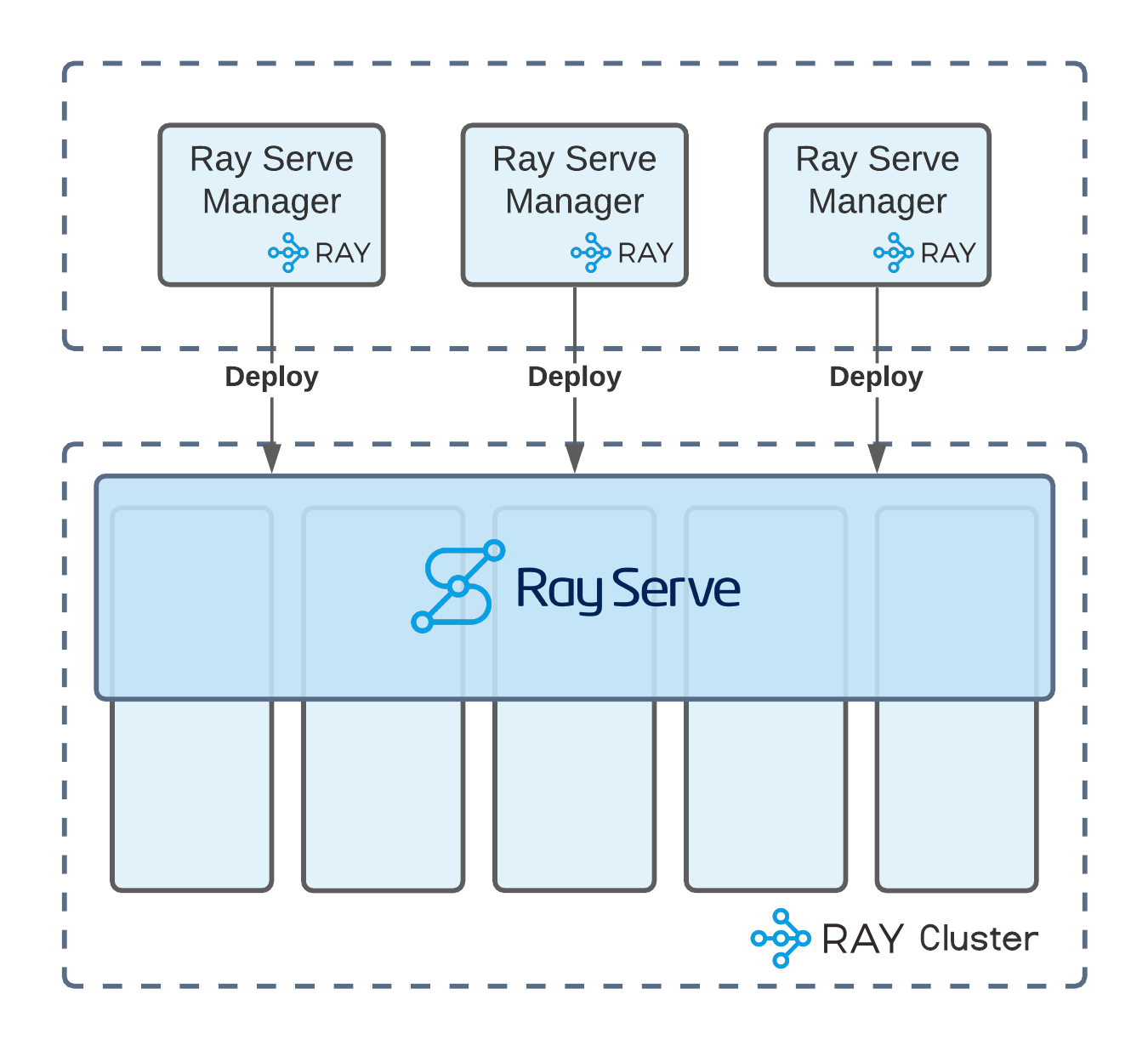
Establishing an interactive data pipeline enabled our users to have faster development cycles with their business logic written in Python. Such fast-paced cycles encouraged more frequent updates of their Python custom scripts, which led to our system re-deploying the same Ray Serve deployments repeatedly. These updates are often executed concurrently as the Ikigai data pipeline allows multiple team members to collaborate even within the same data pipeline. In that scenario, we encountered race condition problems as multiple users tried to update the same Serve instance at the same time. To resolve this challenge, we collaborated with the Ray Serve team at Anyscale to contribute a patch to Ray Serve that allowed us to solve this problem by setting a `prev_version` flag when updating a deployment, allowing us to detect and avoid race conditions.
1# Plain Deployment
2CustomService.deploy()
3
4# Version-aware Deployment
5CustomService.options(
6 version="new_version", prev_version="old_version"
7).deploy()With the version-aware deployment, the Ikigai data pipeline system can now catch unwanted concurrent deployments.
LinkSkewed Traffic on Serve Instance
Unlike other challenges introduced above, we faced a challenge which does not break the system or the mission of the Ikigai data pipeline but is more related to resource management optimization. With the nature of platform users’ unpredictable behavior, a small subset of Serve instances were having highly concentrated traffic. This syndrome showed that if we scale all Serve instances equally, we would suffer from lack of availability for popular Serve instances while wasting resources on unpopular instances. To tackle this problem, we decided to keep track of the amount of traffic on each Serve instance and scale each instance proportionally to its traffic size using Ray Serve’s in-house replica count control.
1@serve.deployment(
2 name="deduplicate_service",
3 num_replicas=2
4)
5class DeduplicateService:
6
7 def deduplicate(self, data):
8 data.drop_duplicates(inplace=True)
9 return data
10
11@serve.deployment(
12 name="sort_service",
13 num_replicas=5
14)
15class SortService:
16
17 def sort(self, data, target_columns, descending):
18 data.sort_values(
19 target_columns,
20 ascending=not descending,
21 inplace=True
22 )
23 return dataThroughout the adoption of Ray Serve to our system, we were fairly impressed by how well-designed the Ray Serve APIs and its deployment managements were. As a software engineer trying to incorporate a new framework to an existing platform, we felt the experience has been very smooth as most of the challenges we faced did not require significant ‘patching’; we could find in-house solutions for our problems following Ray Serve’s design.
LinkFuture of Ikigai Data Pipeline
Having a working system of scalable and interactive data pipeline with Ray Core and Ray Serve, we decided to incorporate a few more Ray sub-projects to take our platform further.
LinkRay Client
Ray Client provides a very simple way to connect a local Python script to a Ray Cluster.
1import pandas as pd
2import ray
3
4# Remotely connect to Ray Cluster
5ray.client("http://ray-client-service.ray:10001").connect()
6
7@serve.deployment(name="custom_service")
8class CustomService:
9
10 def deduplicate(self, data):
11 data.drop_duplicates(inplace=True)
12 return data
13
14df = pd.DataFrame([[1, 2, 3], [1, 2, 3], [2, 3, 4]])
15obj_ref = CustomService.get_handle().deduplicate.remote(data=df)
16result_df = ray.get(df_obj_ref)Adopting Ray Client into the Ikigai data pipeline cluster will be extremely beneficial, as it will help us eliminate the task submission step completely.
LinkRay Workflow
Ray Workflow is yet another Ray sub-project which is under active construction. Ray Workflow aims to add fault-tolerance to any Ray remote tasks and provide recovery of the tasks on distributed systems. Having Ray Workflow embedded as a backbone of the Ikigai data pipeline will not only boost the user experiences by saving their time but also optimize the resource management of the system.
LinkSummary
We believe the Ray team is building a truly awesome product. It is helping Ikigai Labs build a very unique data pipeline system. We are excited to evolve our platform even further, alongside the Ray team’s endeavor.
We talked about this topic at Ray Summit 2021 and you can watch the full video here.
Check out some client solutions we were able to build and if you are interested in trying out our product, book a demo here. We are also hiring!
LinkSpecial Thanks
James Oh, Robert Xin, Robbie Jung @ Ikigai Labs for helping us design the system
Edward Oakes @ Anyscale for introducing Ray Serve to us and his effort of helping us with Ray integration
Yi Cheng, Siyuan (Ryans) Zhuang @ Anyscale for introducing Ray Workflow and detailed guidance
Michael Galarnyk @ Anyscale for helping us organize this post